A team of researchers from Google Research, Google DeepMind, and Yale released C2S-Scale 27B, a 27-billion-parameter foundation model for single-cell analysis built on Gemma-2. The model formalizes single-cell RNA-seq (scRNA-seq) profiles as “cell sentences”—ordered lists of gene symbols—so that a language model can natively parse and reason over cellular states. Beyond benchmarking gains, the research team reports an experimentally validated, context-dependent pathway: CK2 inhibition (silmitasertib/CX-4945) combined with low-dose interferon amplifies antigen presentation, a mechanism that could make “cold” tumors more responsive to immunotherapy. The result is ~50% increase in antigen presentation in vitro under the combined condition.
Understanding the model
C2S-Scale converts a high-dimensional expression vector into text by rank-ordering genes and emitting the top-K symbols as a gene-name sequence. This representation aligns single-cell data with standard LLM toolchains and allows tasks such as cell-type prediction, tissue classification, cluster captioning, perturbation prediction, and biological QA to be phrased as text prompts and completions.
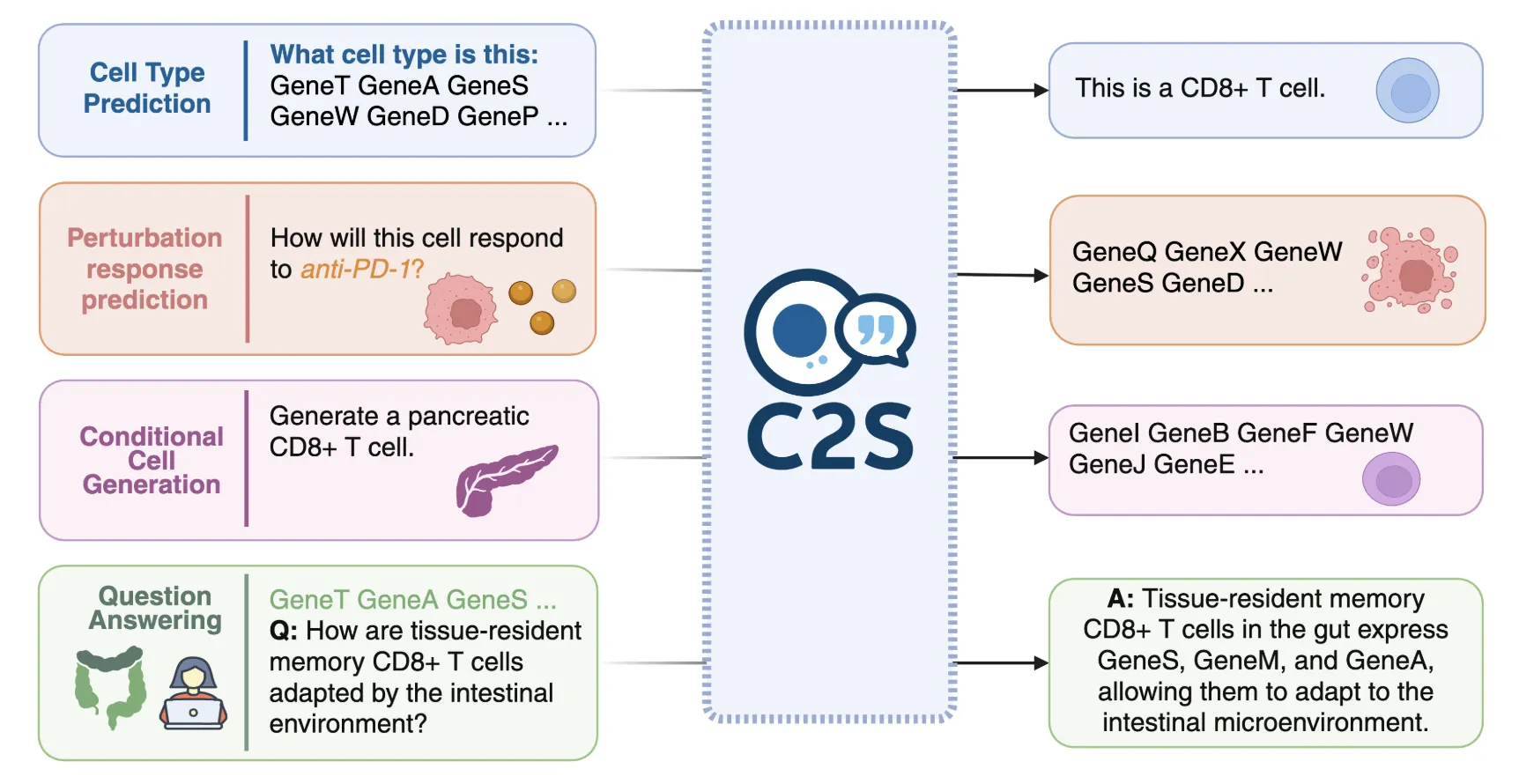
Training data, stack, and release
C2S-Scale-Gemma-2-27B is built on Gemma-2 27B (decoder-only Transformer), trained on Google TPU v5, and released under CC-BY-4.0. The training corpus aggregates >800 public scRNA-seq datasets spanning >57M cells (human and mouse) with associated metadata and textual context; pretraining unifies transcriptomic tokens and biological text into a single multimodal corpus.
The key result: an interferon-conditional amplifier
The research team constructed a dual-context virtual screen over >4,000 drugs to find compounds that boost antigen presentation (MHC-I program) only in immune-context-positive settings—i.e., primary patient samples with low interferon tone—while having negligible effect in immune-context-neutral cell-line data. The model predicted a striking context split for silmitasertib (CK2 inhibitor): strong MHC-I upregulation with low-dose interferon, little to none without interferon. The research team reports in-lab validation in human neuroendocrine models unseen in training, with the combination (silmitasertib + low-dose interferon) producing a marked, synergistic increase in antigen presentation (≈50% in their assays).
The amplifier lowers the response threshold to interferon rather than initiating antigen presentation de novo; flow-cytometry readouts show HLA-A,B,C upregulation only under combined treatment (including IFN-β and IFN-γ), across two neuroendocrine models, with representative MFI gains (e.g., 13.6% @10 nM and 34.9% @1000 nM silmitasertib in one model).
Key Takeaways
- C2S-Scale 27B (Gemma-2) encodes scRNA-seq profiles as textual “cell sentences,” enabling LLM-native single-cell analysis workflows.
- In a two-context virtual screen (>4,000 compounds), the model predicted an interferon-conditional amplifier: CK2 inhibition (silmitasertib) boosts MHC-I antigen-presentation only with low-dose IFN.
- Wet-lab tests in human neuroendocrine cell models confirmed the prediction, with ~50% antigen-presentation increase for silmitasertib+IFN versus either alone; this remains preclinical/in vitro.
- Open weights and usage docs are live on Hugging Face (vandijklab) with both 27B and 2B Gemma variants for research use.
C2S-Scale 27B is a technically credible step for LLMs in biology: translating scRNA-seq into “cell sentences” lets a Gemma-2 model run programmatic queries over cell states and perturbations, and in practice it surfaced an interferon-conditional amplifier—silmitasertib (CK2 inhibition)—that increases MHC-I antigen presentation only with low-dose IFN, a mechanism the team then validated in vitro. The value here isn’t headline rhetoric but the workflow: text-native screening across >4k compounds under dual immune contexts to propose a context-dependent pathway that may convert immune-“cold” tumors toward visibility. That said, all evidence is preclinical and bench-scale; the right read is “hypothesis-generating AI” with open weights enabling replication and stress-testing, not a clinical claim.
Check out the Technical Paper, Model on HF, GitHub Page and Technical details . Feel free to check out our GitHub Page for Tutorials, Codes and Notebooks. Also, feel free to follow us on Twitter and don’t forget to join our 100k+ ML SubReddit and Subscribe to our Newsletter. Wait! are you on telegram? now you can join us on telegram as well.
Michal Sutter
Michal Sutter is a data science professional with a Master of Science in Data Science from the University of Padova. With a solid foundation in statistical analysis, machine learning, and data engineering, Michal excels at transforming complex datasets into actionable insights.


